Different types of PCR techniques in molecular biology
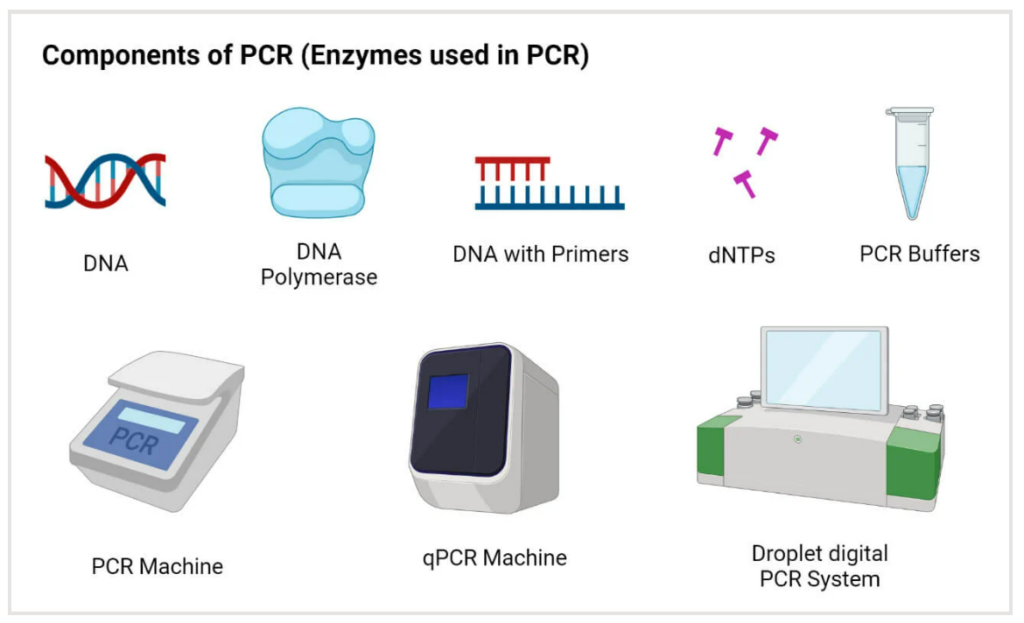
Polymerase chain reaction (PCR) technology is a fundamental tool in molecular biology to rapidly amplify DNA sequences. PCR technique is based on the principle of enzymatic replication of nucleic acids and is one of the main methods for DNA analysis. It was developed in 1983 by the American biochemist Kary Mullis for which he was also awarded the Nobel prize in Chemistry in 1993 together with Michael Smith (1).
PCR has a wide range of applications in cloning, genotyping, mutation detection, sequencing, forensics, and paternity testing (2). Different types of PCR processes with minor modifications can be used to produce better results. The article gives an overview of the commonly used PCR techniques (3).
1. Standard PCR: Standard PCR is the simplest, most efficient, and most sensitive technique to amplify DNA segments using a pair of primers that flank a specific DNA region. Standard PCR consists of three major steps to amplify the target DNA segments: denaturation, annealing, and extension.
2. Reverse transcriptase PCR (RT-PCR): RT-PCR first converts RNA to complementary DNA (cDNA) using the reverse transcriptase enzyme. The cDNA is further amplified using PCR.
3. Nested PCR: Nested PCR is a method to improve sensitivity and specificity involving two primer sets and two successive PCR reactions. The first set of primers anneal to sequences upstream from the second set of primers and the product of the first amplification is used as the template for the second PCR (4).
4. Real-time PCR: In real-time PCR, the accumulation of amplified products is measured while the reaction is progressing. On the contrary, conventional PCR only allows amplicon detection at the end of the reaction. Real-time PCR is an advanced way to quantify DNA or RNA by incorporating a fluorescent probe into the reaction.
5. Multiplex PCR: a multiplex PCR is used to amplify and target multiple DNA sequences in a single reaction well. This variation of PCR requires two or more probes that can be distinguished from each other and detected simultaneously (5).
References
1. Kaunitz JD. The Discovery of PCR: ProCuRement of Divine Power. Dig Dis Sci. 2015 Aug;60(8):2230-1. doi: 10.1007/s10620-015-3747-0. PMID: 26077976; PMCID: PMC4501591.
2. Lenstra JA. The applications of the polymerase chain reaction in the life sciences. Cell Mol Biol (Noisy-le-grand). 1995 Jul;41(5):603-14. PMID: 7580841.
3. Gupta N. DNA Extraction and Polymerase Chain Reaction. J Cytol. 2019 Apr-Jun;36(2):116-117. doi: 10.4103/JOC.JOC_110_18. PMID: 30992648; PMCID: PMC6425773.
4. Green MR, Sambrook J. Nested Polymerase Chain Reaction (PCR). Cold Spring Harb Protoc. 2019 Feb 1;2019(2). doi: 10.1101/pdb.prot095182. PMID: 30710024.
5. Elnifro EM, Ashshi AM, Cooper RJ, Klapper PE. Multiplex PCR: optimization and application in diagnostic virology. Clin Microbiol Rev. 2000 Oct;13(4):559-70. doi: 10.1128/CMR.13.4.559. PMID: 11023957; PMCID: PMC88949.
